The Latest GTN News
Keep an eye on this page for the latest news around the GTN. New tutorials, GTN features, upcoming training events, and much much more!
Want to add your own news here (e.g. new tutorial, event, publication, anything else training related)? Check out how to do that in this FAQ

GTN is now integrated with WorkflowHub
2 September 2025
Thanks to a collaborative effort between the teams at Galaxy Training Network (GTN), WorkflowHub, and Australian BioCommons, GTN workflows are now registered automatically with WorkflowHub. The existing set of workflows can be viewed here: GTN on WorkflowHub. For every new tutorial that is added to the GTN, any workflows that it contains will now also be pushed to WorkflowHub.

Quantifying Electrophoresis Gel Bands with QuPath and Galaxy
18 August 2025
Gel Electrophoresis is a staple of molecular biology, providing a way to separate DNA, RNA, or proteins by size and charge. While gels are usually interpreted visually, the ability to quantify band intensities offers much richer insights—allowing researchers to estimate product yields, verify PCR amplification, or assess protein purity.

Scaling Up Hands-On Bioinformatics Training with TIAAS – An Open University Perspective
3 July 2025
Back in September 2024, we ran the Open University Bioinformatics Bootcamp—a free, five-day online course introducing students to the core tools and techniques used in single-cell biology. We were genuinely delighted by the level of interest: 120 students signed up, 100 showed up, and around 80 worked through the hands-on tutorials during the week. That’s a fantastic level of engagement, especially for a course that’s entirely optional and doesn’t count towards their degree.
Enhancing Scientific Training: The Galaxy Training Network's Role in the ELIXIR Training Life-Cycle
1 July 2025
In the rapidly evolving landscape of data science, continuous learning and skill development are crucial. The Galaxy Training Network (GTN) plays a pivotal role in this educational ecosystem, particularly within the ELIXIR Training Life-Cycle. This blog post explores how the GTN contributes to each phase of the life-cycle and aligns with the SPLASH recommendations, ensuring high-quality training for researchers worldwide.

Galaxy meets Onedata — distributed storage for science
17 March 2025
Thanks to the efforts undertaken in the
EuroScienceGateway project, Galaxy
now offers integration with Onedata. It can be used as
a remote source for data import/export (a.k.a. Files Source Plugin) and as
a storage location for Galaxy datasets (an Object Store).
The integration includes
BYOD
(Bring Your Own Data) and
BYOS
(Bring Your Own Storage) templates.

An Ode to Helena - from the 🥓Bacon Brigade
7 March 2025
@gtn:hexylena, as a key member of the phenomenally creative and passionate Galaxy Training Network team alongside @gtn:shiltemann and @gtn:bebatut, pioneered the concept of Training Infrastructure. In a world dominated by clunky virtual learning environments and expensive software, she built an open-source system that could seamlessly support individual users, trainers, and contributors across diverse disciplines and expertise levels.
Credit where it's due: GTN Reviewers in the spotlight
2 December 2024
We would like to recognise and thank all of the reviewers who have contributed to the GTN tutorials; your efforts are greatly appreciated, and we are grateful for your contributions to the GTN community. Today, we are highlighting your efforts on every single learning material across the GTN.
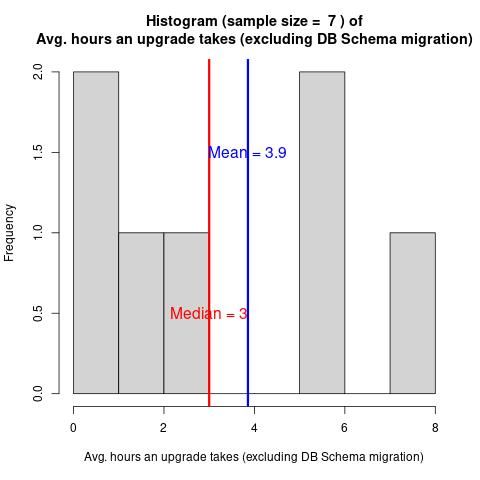
Galaxy Administrator Time Burden and Technology Usage
22 July 2024
Have you wondered how difficult Galaxy is to run? How much time people must spend to run Galaxy?
In February 2024, we collected 9 responses from the Galaxy Small Scale Admin group.
The questions cover various time burdens and technological choices.
The report provides answers to prospective future admins’ most common questions.
Registration Open for BioNT Community Event & CarpentryConnect Heidelberg
11 July 2024
Are you interested in training and want to connect with other enthusiastic trainers? Then join the BioNT Community Event & CarpentryConnect - Heidelberg 2024. The Carpentries and other learning communities will meet to network and collaborate during this 3-day event. The GTN will be present with a Poster, Lightning Talks and a Mini-hackathon.

4th Mycobacterium tuberculosis complex NGS made easy
8 July 2024
Tuberculosis (TB) is a big killer in many countries of the world, particularly in those with low and middle income. Next-generation sequencing has been key in improving our understanding of drug resistance acquisition and of transmission of Mycobacterium tuberculosis. Yet, the need for expertise guiding NGS implementation in laboratories and the lack of bioinformatic expertise, are main obstacles hindering the implementation of NGS into TB programs.
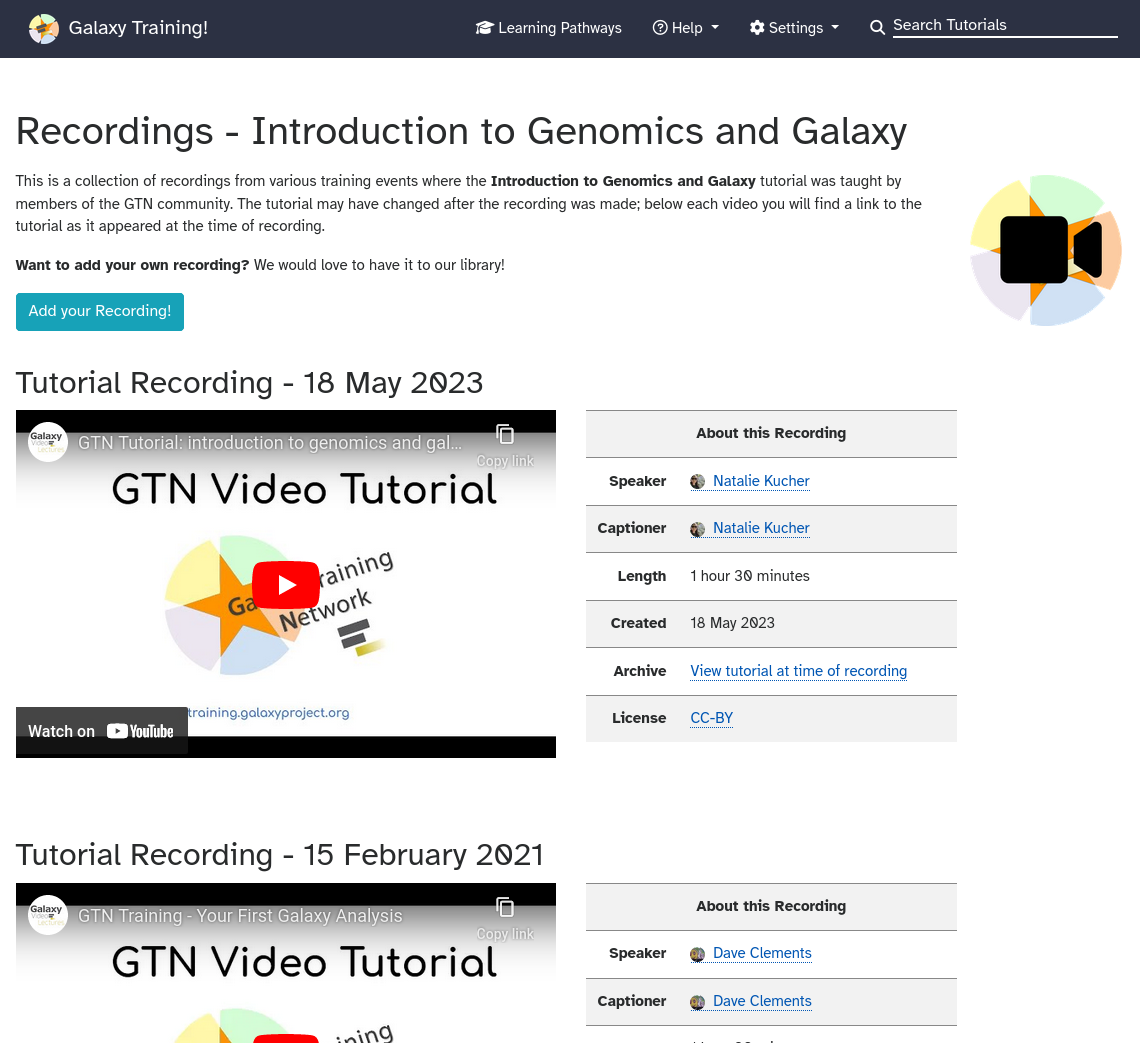
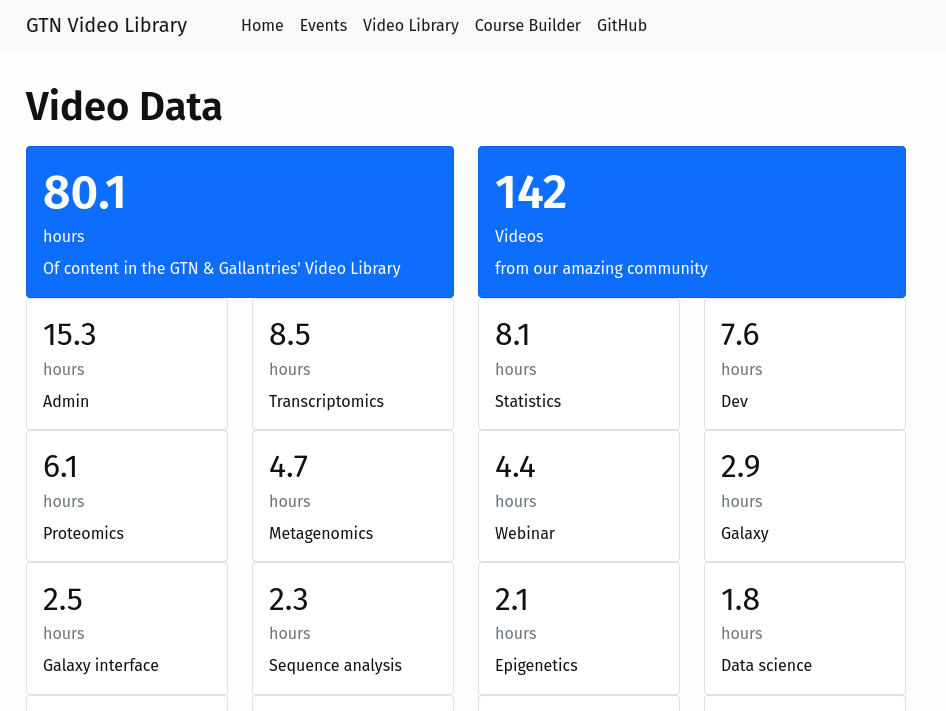
GTN Video Library 2.0: 107 hours of learning across 154 videos
14 June 2024
Many GTN tutorials already have recordings. These recordings were made by members of the community for a variety of (online) training events.
Up until now, this video library were part of the Gallantries Project.
We have now integrated this video library directly into the GTN, and made it even easier to add video recordings to GTN tutorials or slide decks! Just use a Google Form to submit your video recordings!
Phylogenetics tutorial takes researchers back to basics!
13 June 2024
A new Galaxy Training Network tutorial has been created to take researchers back to basics to uncover the principles of phylogenetics and how tree-building methods work. A longstanding collaboration between Professor Michael Charleston from the University of Tasmania and Australian BioCommons has delivered this self-guided tutorial featuring videos and hands-on exercises. To maximise its impact, the resource was tailored specifically to be shared globally via the Galaxy Training Network, and will form the basis of an upcoming live training workshop.

From GTN Intern to Tutorial Author to Bioinformatician
13 June 2024
With growing access and interest in sequencing data, Galaxy is a knight in shining armor for wet lab scientists hoping to analyze their own data. With long term intentions of increasing access to bioinformatic analyses, the Galaxy Training Network (GTN) creates a safe space where non-computer-scientists may analyze their own data and even learn to code: an invaluable skill in today’s scientific world. Galaxy introduced me to brand new skills as an undergraduate and ultimately changed the trajectory of my career. Here is my story as a biology undergraduate with no coding experience turned GTN contributor &, eventually, coding bioinformatician: thanks to Galaxy.
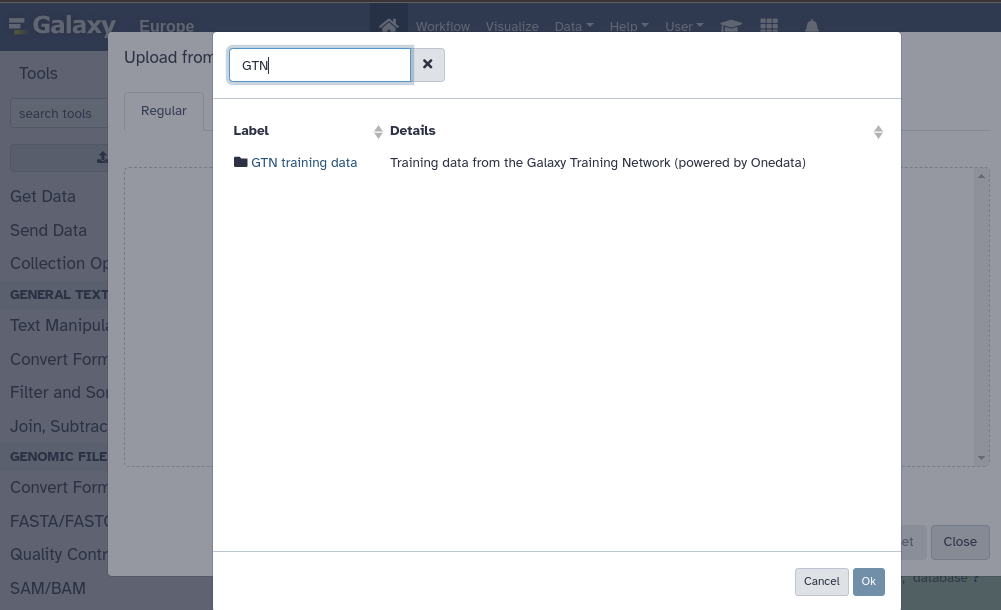
All GTN training data are now automatically mirrored via Onedata
3 June 2024
As part of the EuroScienceGateway and in cooperation with Onedata and EGI we are providing all GTN training data on a publicly accessible cloud storage.
Those training datasets are curated, small but meaningful for educational purposes and contain 1530 files with a total size of 170Gb.
An invaluable set of resources for everyone dealing with data science and training.
Please thank the more than 350 contributors to GTN.
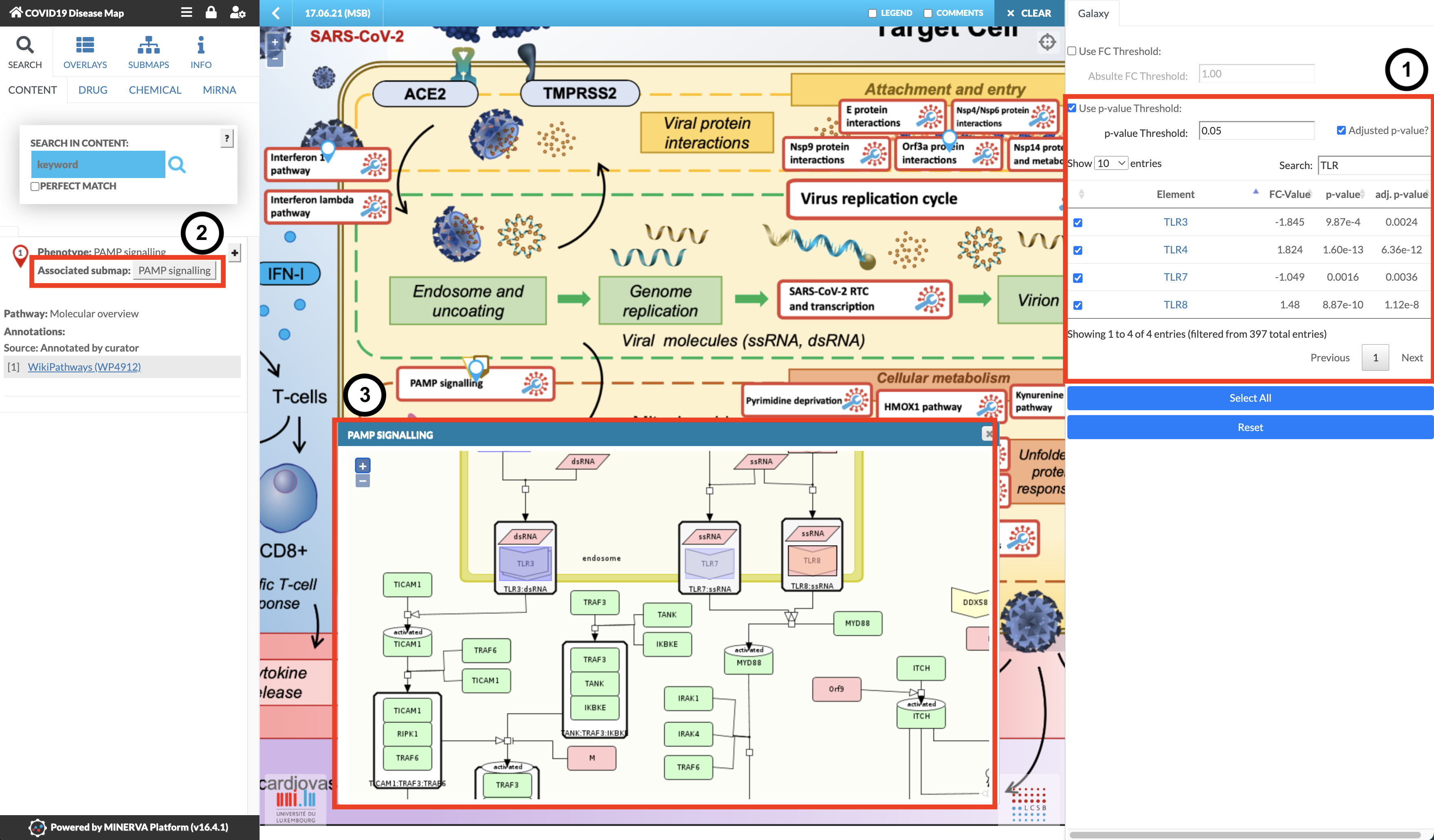
Learn to use MINERVA Platform's COVID-19 Disease Map with Galaxy
28 March 2024
As part of the work of BeYond COVID, we have developed a new integration between the MINERVA Platform’s COVID-19 Disease Map and Galaxy! Datasets created within Galaxy can now be seamlessly visualized in the MINERVA Platform, allowing you to explore your data in the context of the COVID-19 Disease Map.

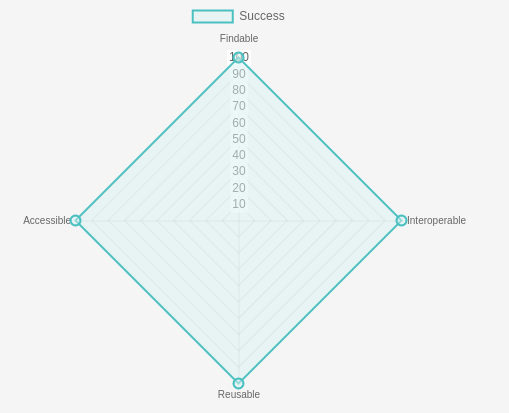
Cool URLs Don't Change, GTN URLs don't either.
18 March 2024
At the Galaxy Training Network we are really committed to ensuring our training materials are Findable, Accessible, Interoperable, and Reusable.
This means that we want to make sure that the URLs to our training materials are persistent and don’t change.
The GTN wants you to be able to rely on our URLs once you’ve added them to a poster or training material, without having to worry about them breaking in the future.

GTN in Discourse
30 January 2024
Howdy teachers and support staff! We are exploring displaying GTN content directly in the Galaxy Help site, furthering the integration between the GTN and the Galaxy community infrastructure.
When you paste a GTN Frequently Asked Questions URL into the Galaxy Discourse, it will now be rendered directly in Discourse!

Galaxy Single-cell Community: Year in Review
22 December 2023
🚀Embarking on a cosmic journey, the Galaxy Single-cell Community has clustered together to unveil a constellation of tools, making strides in RNA-stellar discoveries and creating out-of-this-world workflows. With a commitment to battling work duplication across the multiverse, this community is boldly charting a course for global domination, proving that when it comes to bioinformatics, the Galaxy is the limit!✨

User story: Where Galaxy meets medicinal chemistry
18 December 2023
As a medicinal chemistry student, I undertook a semester project on natural products isolation and derivatisation. One of the compounds my group was working on was piperine, extracted from black pepper. Inspired by literature findings, we found out that the derivatives of piperine can inhibit monoamine oxidase-B and thus can be possibly used in Parkinson’s disease. As a novelty element in our project, we came up with new structures of derivatives that could act as inhibitors.
Single cell subdomain re-launch: Unified and feedback-driven
12 October 2023
The European Galaxy Days 2023 CoFest combined the forces of administrator, developer and trainer to update and re-launch the single cell Galaxy instance. Where previously there were two subdomains each with their own sets of tools, there is now a unified subdomain with re-categorized tools that makes sense for users.

ELIXIR-UK Fellow Launches New FAIR Data Management Training
21 June 2023
Katarzyna Kamieniecka (University of Bradford), a member of the ELIXIR-UK Fellowship Programme, has just launched a new series of lessons to improve {FAIR} data management in the life sciences. The lessons are designed to help researchers and research support staff understand the principles of FAIR data management and apply them to their own work.

New Feature: Click-to-run Workflows
19 April 2023
The GTN has implemented “click to run” workflows! One click, will get you into Galaxy with a workflow imported and ready to run.
If you are on either usegalaxy.org or usegalaxy.eu, we have implemented easy buttons which you can click that will direct you straight to your Galaxy server.

New Feature: Trainer Directory! (Add yourself today!)
21 March 2023
Numerous other training groups keep “Trainer Directories” as a way to help students meet trainers in their area, especially for hosting workshops for their local community. There have been a handful of attempts at this in the past, both in galaxyproject/galaxy-maps and in a Google Map on the GTN homepage.

What are the most used tools in the GTN?
8 February 2023
Did you ever wonder which was the number one most used tool in the GTN? There is a new page of Top Tools where we list all of the top tools in the GTN, and which tutorials use them. We guessed “upload1”, but it seems like not every author annotates that step explicitly in their tutorial, and of course it doesn’t appear in the workflows which we also scan to create this tool listing.

GTN Celebrates Black History Month
2 February 2023
February marks Black History Month in the United States. This year we want to celebrate that by introducing a new theme (Click to Activate), and taking a look at one of the most famous Black figures in Science: Henrietta Lacks whose contribution to science went unremarked and unappreciated for decades while white scientists took credit for advancements built on her cell line.
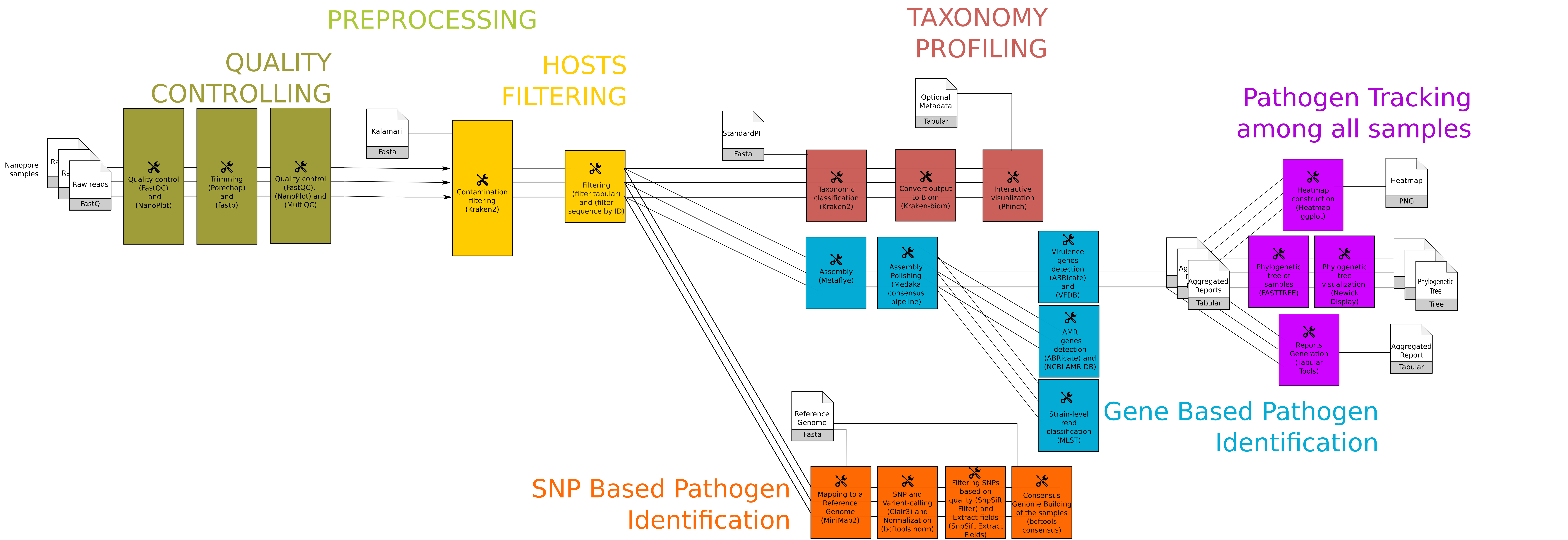
New Tutorial: Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition
26 January 2023
Food contamination with pathogens are a major burden on our society. Globally, they affect an estimated 600 million people a year and impact socioeconomic development at different levels. These outbreaks are mainly due to Salmonella spp. followed by Campylobacter spp. and Noroviruses.

New Feature: Prometheus Metrics endpoint
24 January 2023
Prometheus is an increasingly popular monitoring solution whereby sites expose a /metrics endpoint, and then a central Prometheus server can scrape it for data which is then plotted or visualised or alerted upon. We at the GTN are now testing out a prometheus endpoint for our completely static site which we’ll use to expose some maybe useful statistics about the GTN that others can then scrape and display in ways that are useful for them.

New GTN Feature Tag-based Topics enables new SARS-CoV-2 topic
23 January 2023
The GTN has long struggled with the rigid hierarchy of a single level of classification: topics. All materials had to be organised into a single topic, even when their contents sometimes spanned multiple topics! We added subtopics a while back to help further organise these tutorials, but that still did not solve the issue of needing to collect multiple learn materials in a single location for people to access.
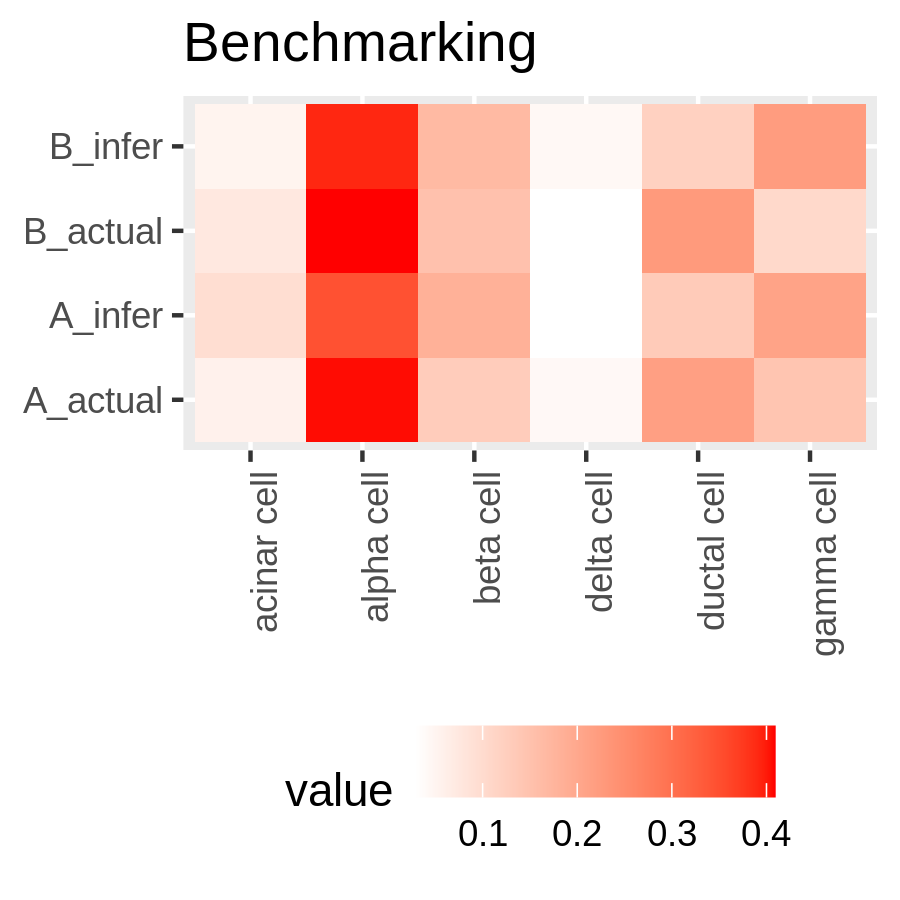
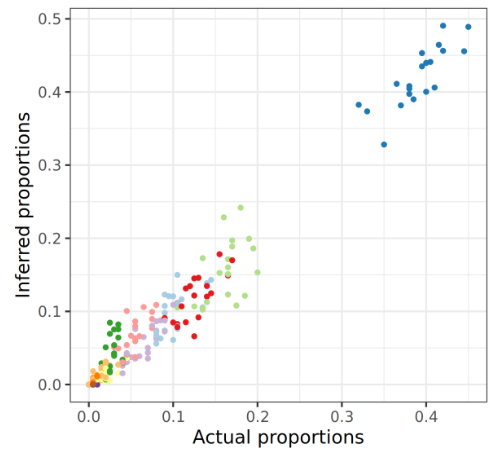
New Tutorial Suite: Deconvolution with MuSiC, from public data to disease interrogation!
29 November 2022
The still new and shiny single-cell analysis topic now boasts a deconvolution tutorial suite! What does deconvolution do you ask? Well, in this context, it infers cell proportions from bulk RNA-seq data. You heard that correctly - instead of expensive new single-cell experiments, you can re-analyse old bulk RNA-seq data and estimate cell proportions. All you need is a reasonably good single cell dataset to use as a reference and you’re good to go! The tutorial suite shows you how to build your reference from publicly available single cell data, and apply analysis to some publicly available bulk RNA-seq data.
New Topic: Single Cell Analysis!
18 November 2022
Single-cell analysis now has it’s own topic! These tutorials were previously part of the transcriptomics topic, but due to the amazing efforts by
Wendi Bacon,
Mehmet Tekmanand others, we now have so many single-cell analysis tutorials that they deserve their own dedicated topic! From introductory slides and practicals, to case study tutorials generating cell clusters and trajectories from raw sequencing files, and even a growing subtopic of smaller tips and tricks for adapting analysis to user needs, the single cell topic either has what you need or is working on it. Come learn or get involved!

New Tutorial: Data Manipulation
11 September 2022
Many analyses in Galaxy require small data manipulations in between tools as a sort of “glue” between analysis steps. For example, you may need to change the file format, re-order columns in a table, replace certain values, or perform some filtering operation, before you can proceed to the next step of your analysis. Galaxy offers a wide range of tools that can perform such generic data manipulation tasks, and mastering these types of operations in Galaxy can help you streamline your analysis and get the most out of Galaxy.
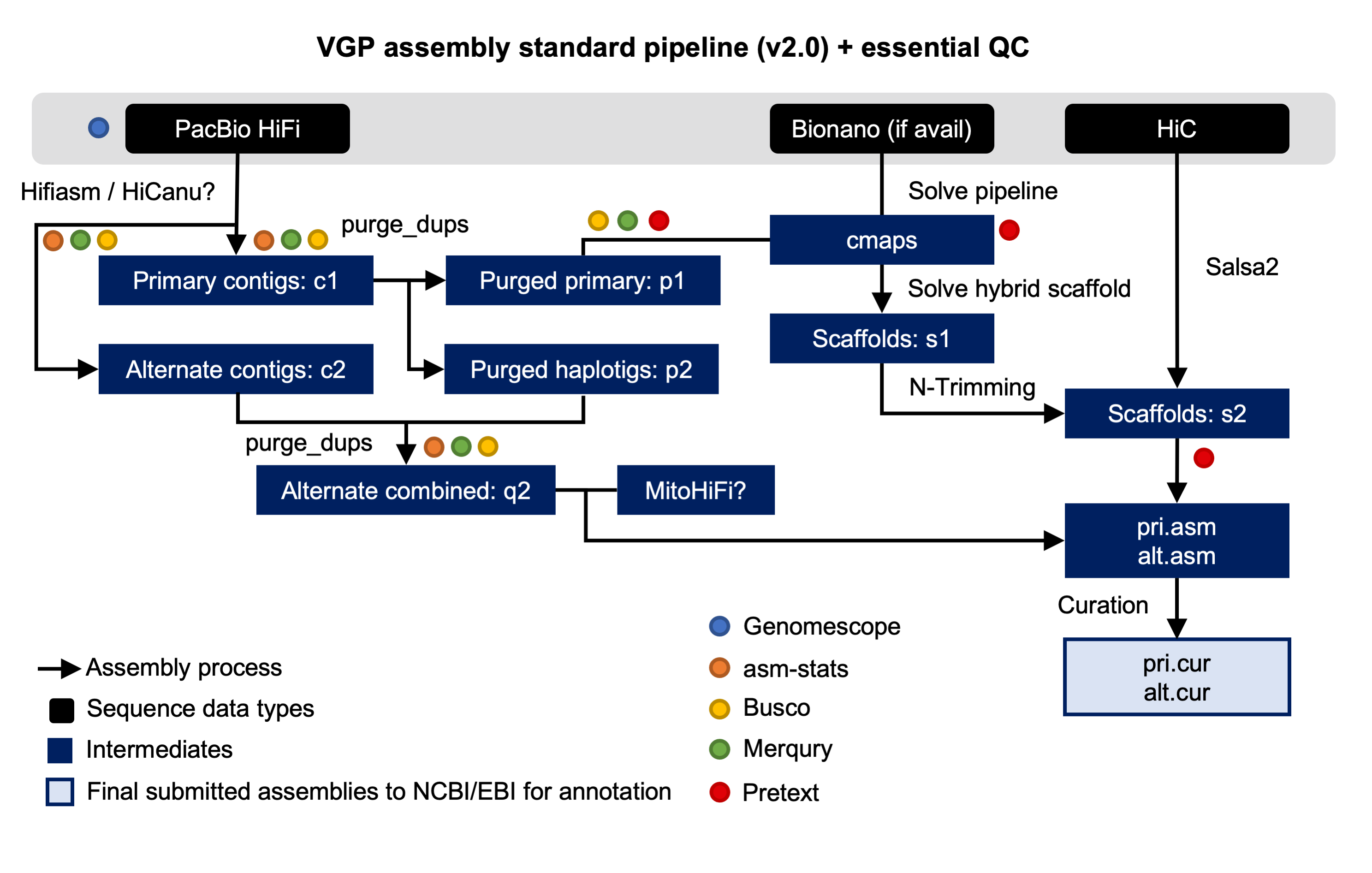
New Tutorial: VGP assembly pipeline
14 March 2022
We are proud to announce that, as result of the collaboration with the Vertebrate Genomes Project (VGP), a new training describing the VGP assembly pipeline is now available in the Galaxy Training Network. The Vertebrate Genomes Project aims to generate high-quality, near-error-free, gap-free, chromosome-level, haplotype-phased, annotated reference genome assemblies for every vertebrate species.
New Feature: Automatic RMarkdown
28 January 2022
Further building on the work in the automatic Jupyter Notebook, we’ve now re-written the Jupyter export to be faster, and more importantly added support for R and RMarkdown! Check out an example material. Here we take the content of the tutorial, written like normal GTN markdown, and we automatically convert it to Jupyter Notebooks and now RMarkdown documents! Check out the documentation on how to setup your tutorials to support this.

New FAQs: How does the GTN stay FAIR and Collaborative
1 December 2021
The GTN does a lot of work to adhere to the best practices for FAIR training materials as set out in Garcia et al. 2020 and a lot of this work is behind-the-scenes or automatic from the infrastructure we provide, and we wanted to highlight that for educators and organisers who want to use GTN materials. This is one of the ways the GTN works to makes itself a fantastic platform training. Find out how contributing simple markdown tutorials can result in fantastically FAIR training.
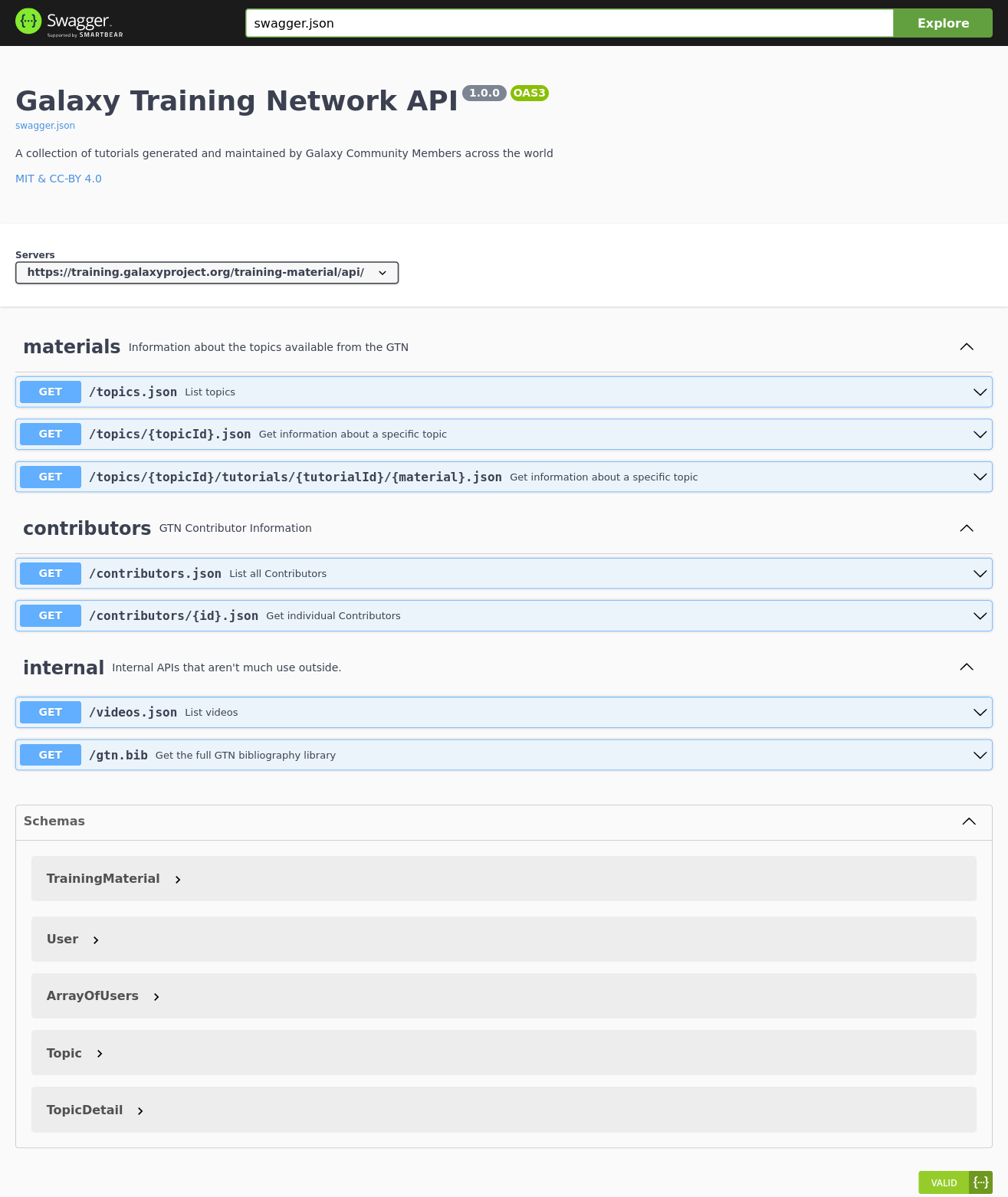
New Feature: GTN API with OpenAPI 3 specification
10 November 2021
As part of the work for the Gallantries Grant, we wanted to access some of the data from the GTN in a more accessible, API based way. We are working on building a Video Library based on the wild success of our Smörgåsbord and GCC2021 events and to support that we wanted to start pulling in data from the GTN to save course authors time and effort when designing their asynchronous courses.
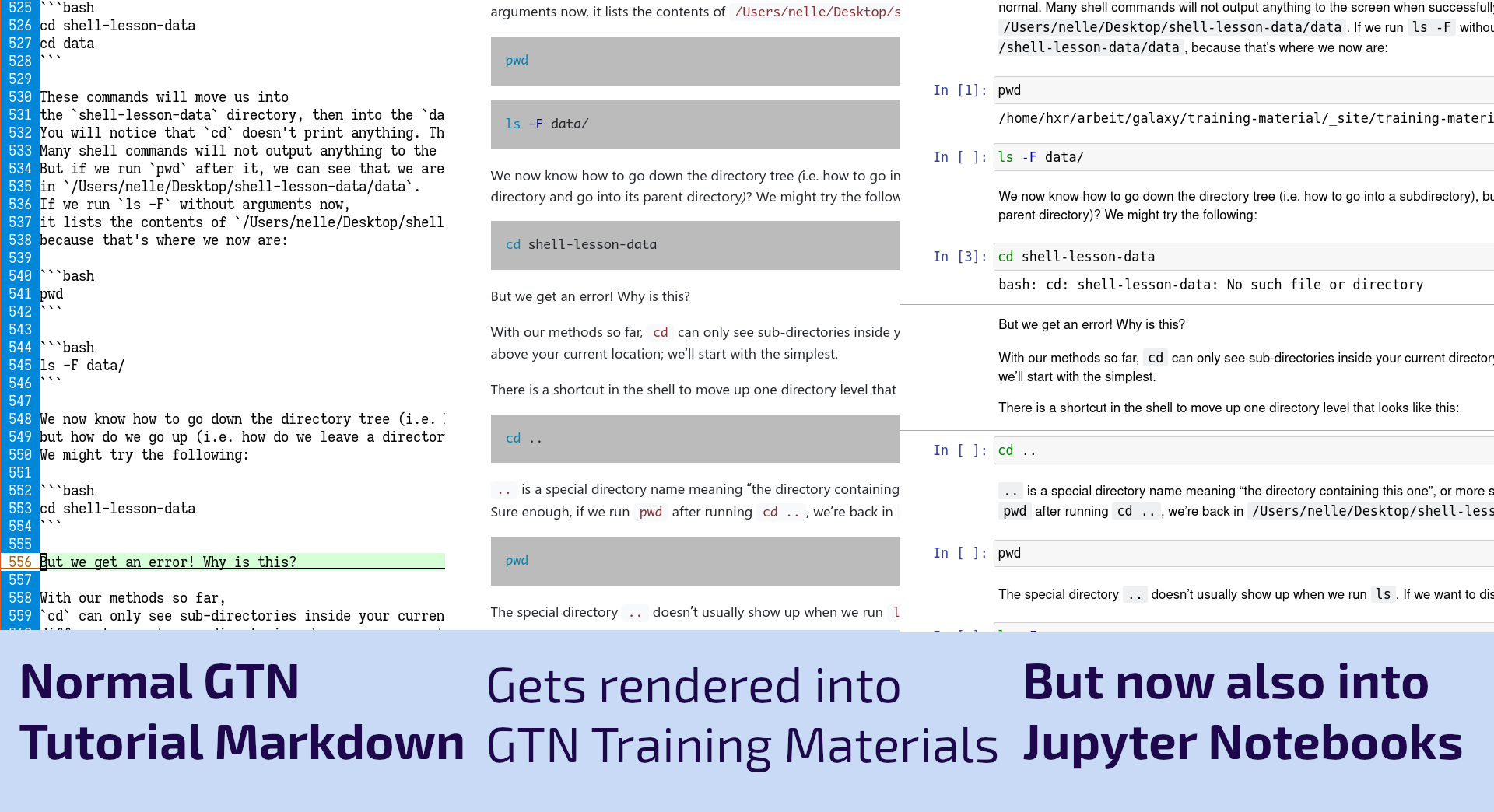
New Feature: Automatic Jupyter Notebooks
24 September 2021
As part of the work for the Gallantries Grant, we found ourselves wanting to teach a number of non-Galaxy tutorials on topics such as Python, Bash, SQL, and Git. Writing the tutorials was always a bit awkward as you would spend a lot of time writing code blocks that would be copied and pasted by students into a terminal. We thought we could do better than that, so we decided we would try to convert these to Jupyter Notebooks where the students could read the training materials and simultaneously execute the code cells directly there with the text! This worked fantastically and now we’ve built this feature into the GTN to make it easier to run all of the coding tutorials.

Accessibility Improvements
27 July 2021
One of our community members suggested on twitter that we support alternative formats for accessing our JavaScript based slides, as this can be difficult for users using screen readers. We’ve now added support for “plain text slides”, where we render the slide decks as a single long document.
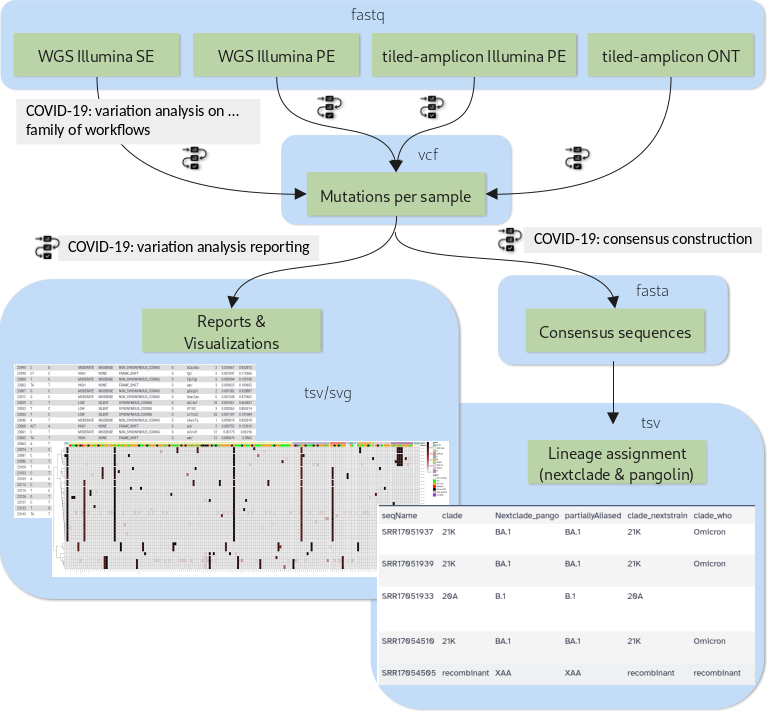
New Tutorial: Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data
30 June 2021
Effectively monitoring global infectious disease crises, such as the COVID-19 pandemic, requires capacity to generate and analyze large volumes of sequencing data in near real time. These data have proven essential for monitoring the emergence and spread of new variants, and for understanding the evolutionary dynamics of the virus.
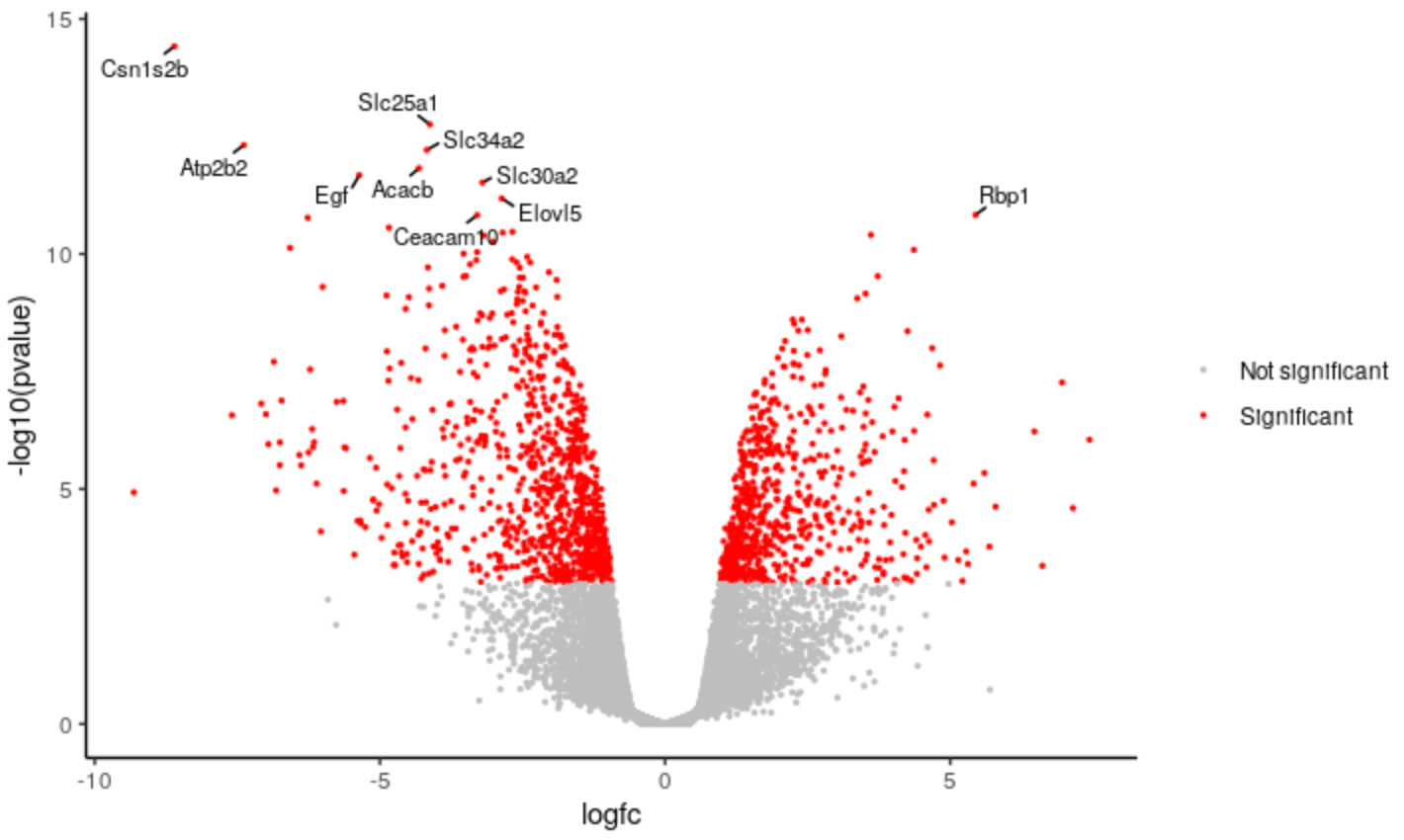
New Tutorial: Visualization of RNA-Seq results with Volcano Plot in R
26 June 2021
The Volcano plot tutorial introduced volcano plots and showed how they can be easily generated with the Galaxy Volcano plot tool. This new tutorial shows how you can customise a plot using the R script output from the tool and RStudio in Galaxy. A short video for the tutorial is also available on YouTube, created for the GCC2021 Training week.
Happy plotting!
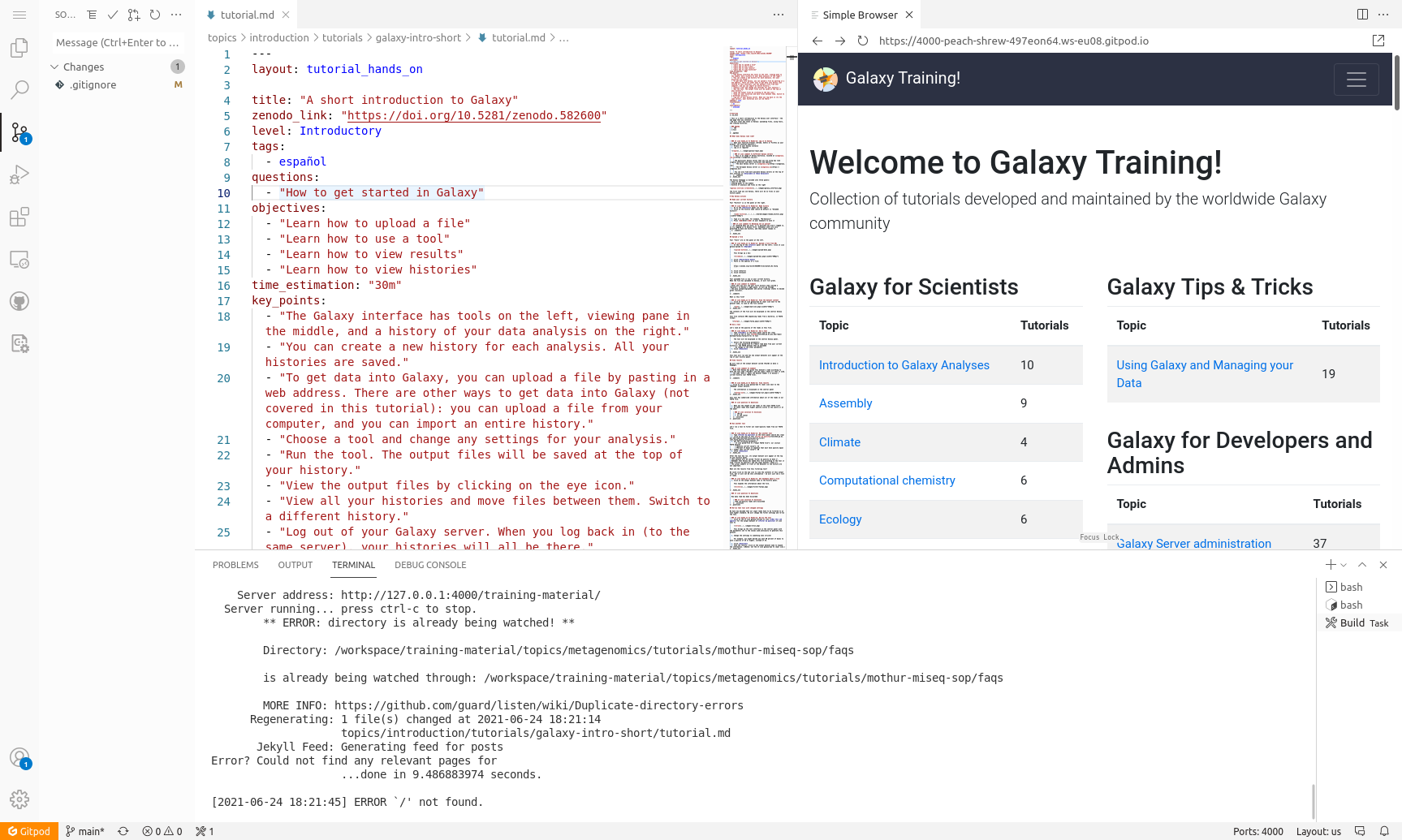
New Tutorial: GitPod for contributing to the GTN
25 June 2021
Want to contribute to the GTN without installing anything on your own computer? GitPod provides an online workspace for editing the GTN tutorials, and showing a live preview of the GTN website with your changes. This tutorial shows you how to set up GitPod, how to use it to get a live preview of the GTN website, and how to save any changes you make back to your GitHub fork. This makes it easier than ever to contribute to the GTN!
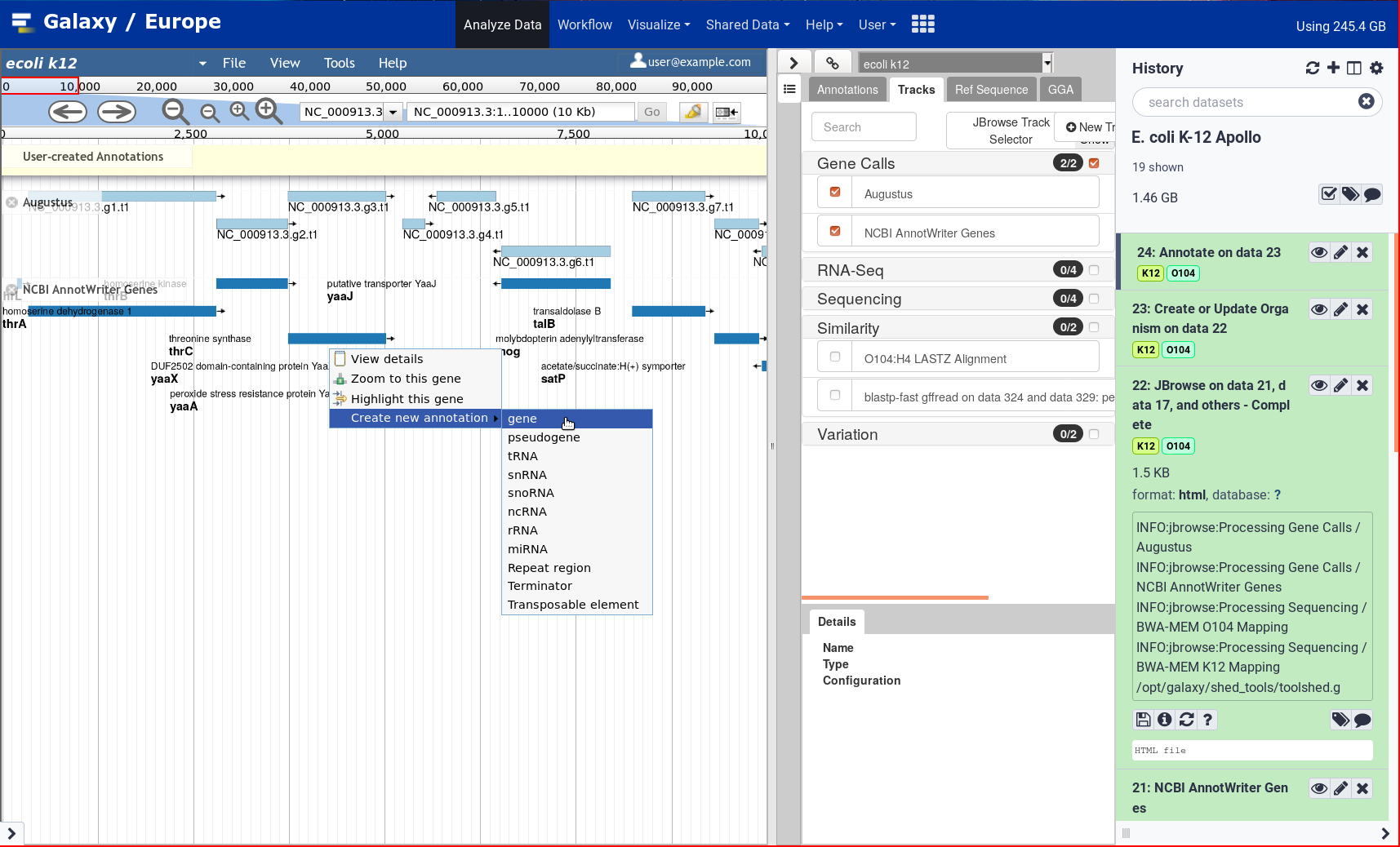
New Tutorial: Genome Annotation with Apollo
4 June 2021
After two years, the Galaxy Genome Annotation team has finally finished their much awaited tutorial on doing Genome Annotation in Galaxy! Genome annotation is a time consuming and largely manual process as it requires the synthesis of a significant amount of varied information to make inferences, a process tha cannot be accomplished without an interactive editor. To support this, Apollo was deployed on UseGalaxy.eu. Over the past two years the GGA project has pushed forward apollo and tooling around it, and it finally culminates in this look at how to annotated your genomes in Apollo.

Oh no, it changed! Quick, to the archive menu.
1 June 2021
Did you ever start teaching a GTN tutorial, only to realise something changed recently? Oh no! A while ago the GTN created an Archive to address just this concern.
On every tag, we’d build a copy of the website which we would place online and never touch again. This way we’d have permanent online copies so that if someone needed to go back to an older version of the GTN, they could.

¿Hablas español?: The first curated tutorial in Spanish!
20 May 2021
¡El primer tutorial en español ya está disponible! Galaxy siempre ha tenido tutoriales traducidos automáticamente mediante Google-Translate, ahora nos estamos embarcando en un nuevo proyecto para estudiar la experiencia de aprendizaje con tutoriales en bioinformática traducidos por humanos. Nuestro objetivo es crear un paquete de tutoriales para el análisis de datos single-cell (célula única) en primera instancia, con el fin de estudiar su uso y utilidad en un taller para hablantes nativos de español. Agradecemos enormemente a las nuevas participantes @gtn:pclo y @gtn:ales-ibt (EMBL-EBI) así como a las miembros existentes de GTN @gtn:beatrizserrano (University of Freiburg), @gtn:shiltemann & @gtn:hexylena (Erasmus-MC), por su arduo trabajo en este proyecto de traducción y mantenimiento y, por supuesto, a @gtn:nomadscientist (The Open University/EBI) como líder de proyecto.
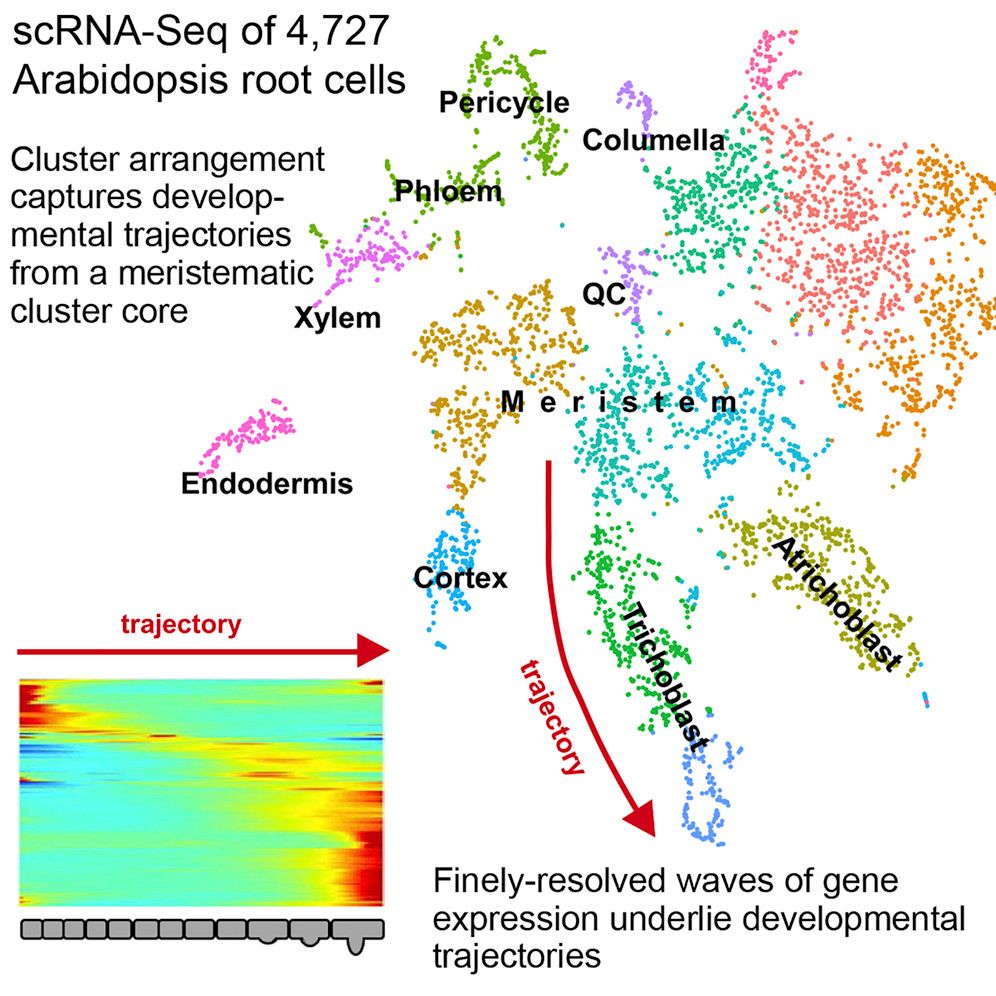
New Tutorials: Whole transcriptome analysis of Arabidopsis thaliana
10 April 2021
Plant lovers are in luck! A new tutorial on this fascinating kingdom has been added to the GTN. It details the necessary steps to identify potential targets of brassinosteroid-induced miRNAs. Brassinosteroids are phytohormones that have the ability to stimulate plant growth and confer resistance against abiotic and biotic stresses. These characteristics have led to great interest in the field of biotechnology due to its potential for increasing agricultural productivity.
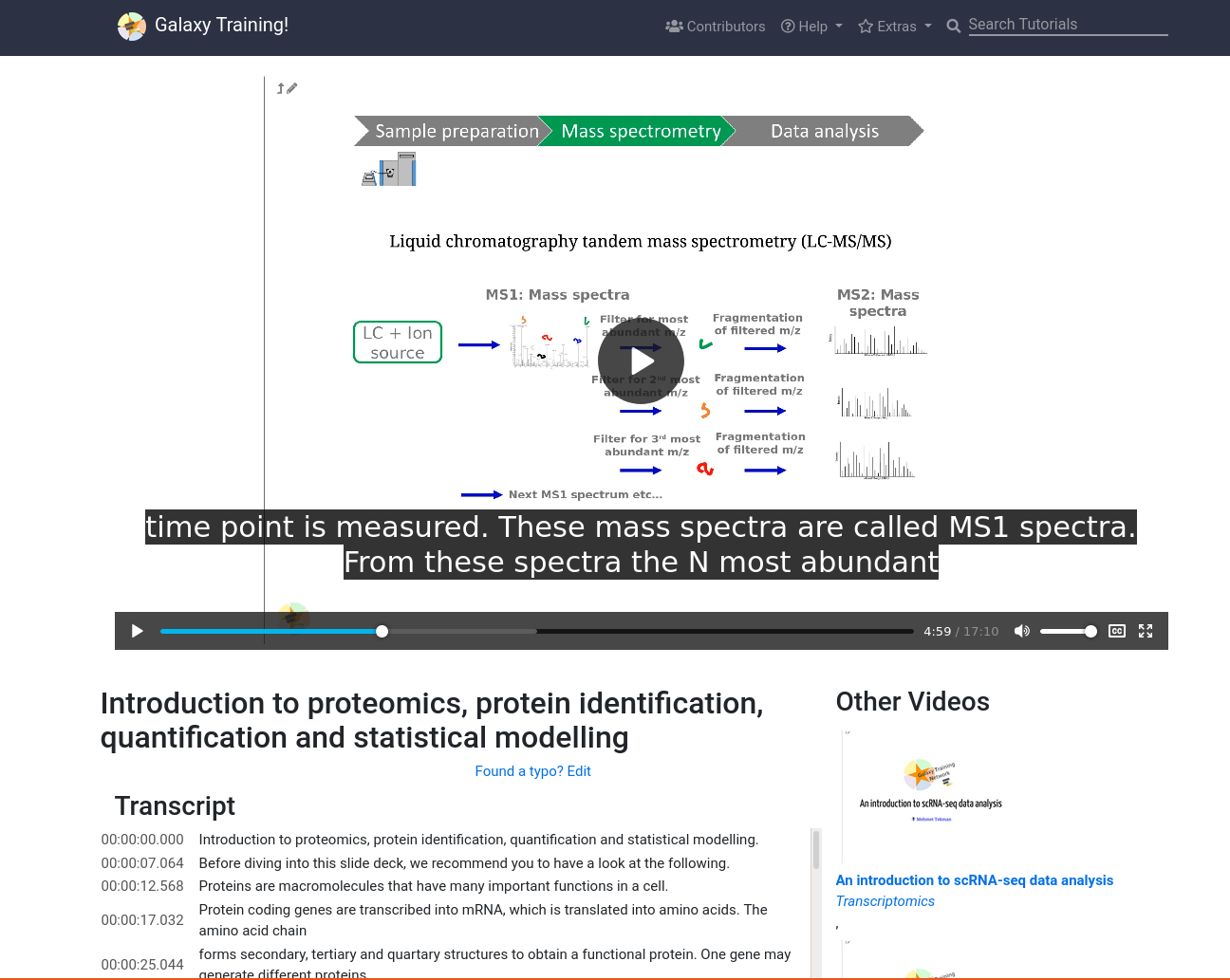
New Feature: Video Player
6 April 2021
One of our contributors (@gtn:mtekman) found himself wanting to link to a specific time point in a video. We said “Wow, what a fantastic idea!” and then we implemented it. To support that we developed a very simple standalone video player page where you can view the GTN videos and browse through our catalog. It features the full transcript on the left, some suggested videos on the right, and a very standard <video> player.

New Tutorials: Genome assembly of a MRSA genome
24 March 2021
Two new assembly tutorials have been added to the GTN! These tutorials walk you through the
process of assembling MRSA strains from real-world datasets. MRSA stands for “Methicillin-
resistant Staphylococcus aureus, it is resistance to several antibiotics and is a common
source of hospital-acquired infections and outbreaks. In this tutorial you will assess the
quality of your data, perform an assembly and assess its quality, analyze your data for the
presence of antimicrobial resistance (AMR) genes, and annotate the genome using Prokka and
JBrowse.

New Feature: FAQs
24 March 2021
Snippets have received an major overhaul! Snippets/FAQs are small reusable bits of training, often answering a single question. In addition to the general Galaxy snippets, we now also support topic-level and tutorial-level FAQs. This will allow contributors to add common questions and answers to their tutorials, which not only can be included in the tutorial text itself, but are also used to autogenerate an FAQ page for the tutorial or topic listing all common questions (and answers).This will be useful both to participants following tutorials via self-study, and also to instructors preparing for a training event.
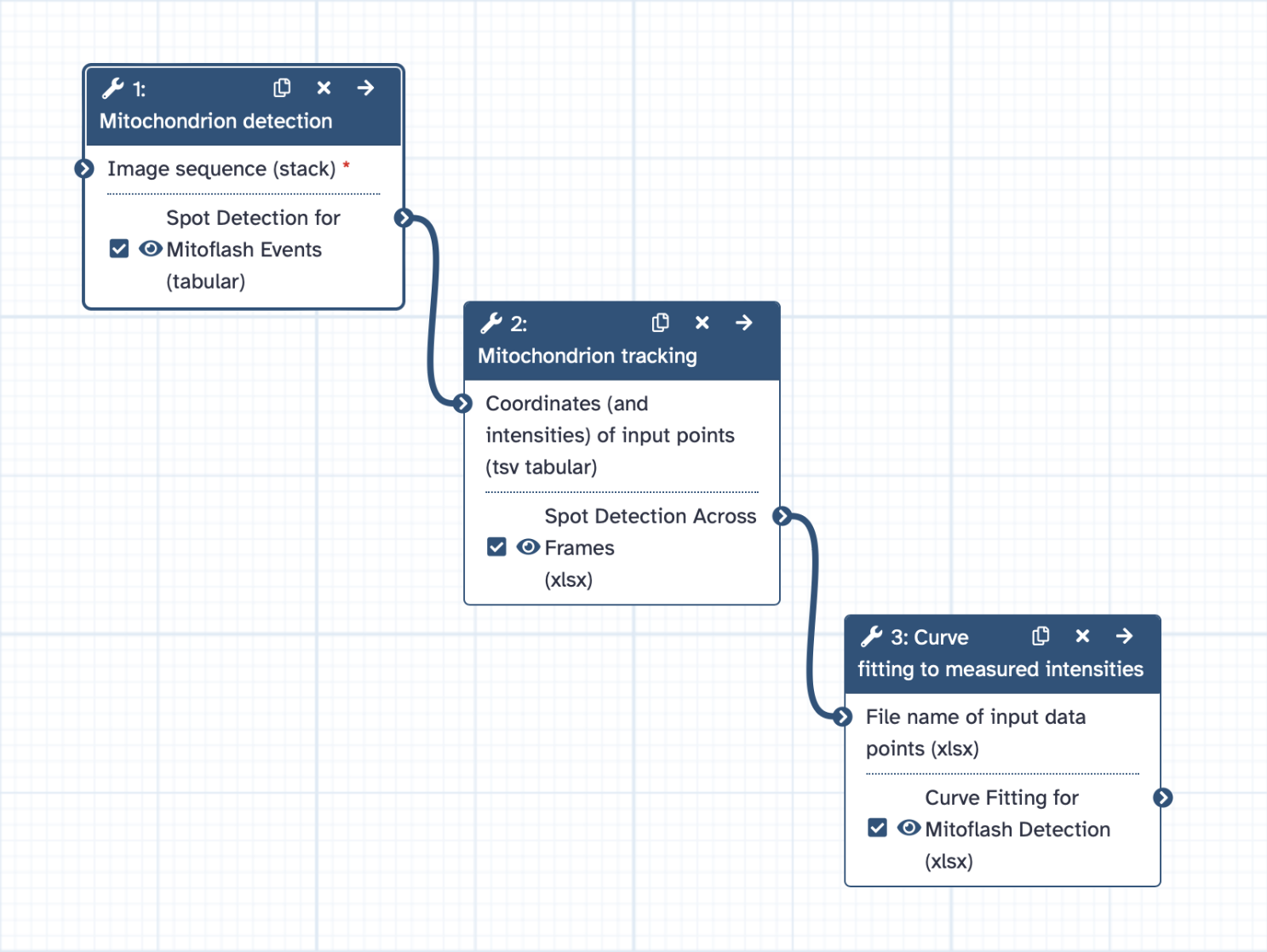
New Tutorial: Object tracking using CellProfiler
18 March 2021
Most biological processes are dynamic and observing them over time can provide valuable insights. Combining fluorescent markers with time-lapse imaging is a common approach to collect data on dynamic cellular processes such as cell division. However, automated time-lapse imaging can produce large amounts of data that can be challenging to process. One of these challenges is the tracking of individual objects as it is often impossible to manually follow a large number of objects over many time points.
Next GTN CoFest May 20, 2021
18 March 2021
Every 3 months we organise one day dedicated to the GTN community! Thursday, May 20th is the next GTN CoFest. We will be working on the training materials, have discussions with the global Galaxy traning community. Do you teach with Galaxy? Want to learn how to add your tutorial to the GTN? New to the community and just want to learn more? Everybody is welcome!