| Author(s) |
|
Posted on: 6 June 2024 purlPURL: https://gxy.io/GTN:N00084
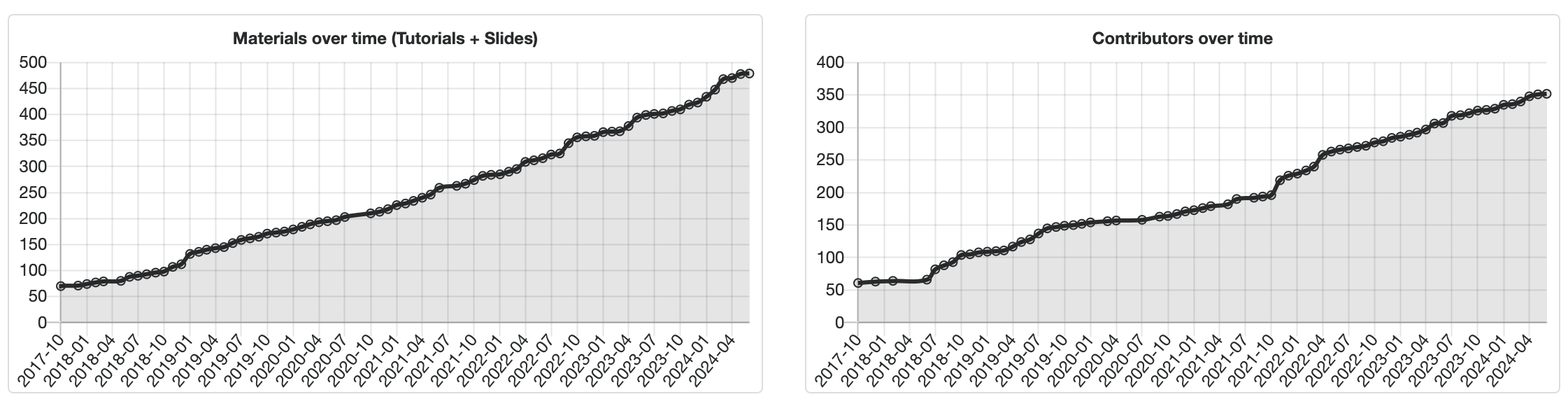
The Galaxy Training Network (GTN) has reached an exciting milestone: our 400th tutorial! This achievement is a testament to the dedication and hard work of our community of educators, researchers, and developers over the last 9 years.
The GTN was established to provide comprehensive and accessible training materials for users of the Galaxy, a widely-used, open-source platform that empowers researchers worldwide to conduct data analysis. Over the years, the network has massively grown, both in content and community engagement, reflecting the dynamic nature of scientific research and the continuous need for up-to-date training resources.
A Journey of Knowledge and Collaboration
From its inception, the GTN has been driven by the goal of making high-quality, user-friendly tutorials available to everyone. The diverse range of tutorials covers topics from basic Galaxy usage to advanced bioinformatics analyses, catering to both beginners and seasoned researchers.
This 400th tutorial represents more than just a number; it embodies the collaborative spirit and the shared pursuit of knowledge that defines the Galaxy community. Each tutorial was contributed by experts who not only have a deep understanding of their subject matter but also a passion for teaching and community building.
Highlights of Our Tutorials
Our tutorials span a wide array of scientific domains and technical skills (30+ topics), including but not limited to:
- Genomics: Explore the fundamentals of sequence analysis, variant calling, and genome assembly.
- Proteomics: Dive into protein identification, quantification, and functional analysis.
- Ecology: Learn to analyse Ecological data through Galaxy.
- Machine Learning: Understand the integration of machine learning techniques in biological data analysis.
- Data Visualization: Master the art of presenting your data in clear and informative ways.
Each tutorial is designed to be interactive, with hands-on exercises that allow users to apply what they’ve learned in real-world scenarios. Moreover, the tutorials are continuously updated to incorporate the latest advancements and tools in the field.
The Power of Community
The GTN’s success is a direct result of the vibrant and collaborative Galaxy community. Contributors from around the globe share their expertise, review content, and provide feedback to ensure the tutorials remain accurate and relevant. This collective effort has created a rich repository of knowledge that is freely accessible to anyone, anywhere. We thank everyone who contributed!
Looking Ahead
As we celebrate this milestone, we remain committed to expanding and enhancing our training materials. We have recently expanded beyond Galaxy into Data Science and command line analyses, and we look forward to expanding to even more new life science topics going forward.
Here’s to the next 400 tutorials and beyond—together, we are advancing science and education, one tutorial at a time!